Extinction catastrophe of antiviral drugs Molnipiravir ( MK-4482)
Extinction catastrophe of antiviral drugs Molnipiravir ( MK-4482), Remdesivir and 4 -fluor uridine (4FIU) acting as a nucleoside analogues against pandemic human coronaviruses.
Juan Carlos López Corbalán, Francisco Marco del Castillo and José Miguel Seguí Ripoll
Extinction catastrophe can be experimentally induced by promutagenic nucleosides in cell culture models. The loss of HIV infectivity has been observed after passage in 5-hydroxydeoxycytidine or 5,6-dihydro-5-aza-20-deoxycytidine while producing a two-fold increase in the viral mutation frequency. Among approved nucleoside analogs, experiments with polioviruses and other RNA viruses suggested that ribavirin can be mutagenic, although its mechanism of action is not clear. Favipiravir and molnupiravir exert an antiviral effect through lethal mutagenesis. Both drugs are broad-spectrum antiviral agents active against RNA viruses. Favipiravir incorporates into viral RNA, affecting the G→A and C→U transition rates. Molnupiravir (a prodrug of β-D-N4-hydroxycytidine) has been recently approved for the treatment of SARS-CoV-2 infection. Its triphosphate derivative can be incorporated into viral RNA and extended by the coronavirus RNApolymerase. Incorrect base pairing and inefficient extension by the polymerase promote mutagenesis by increasing the G→A and C→U transition frequencies. Molnupiravir is the isopropylester prodrug of the ribonucleoside analogue β-D-N4-hydroxycytidine (NHC). NHC is abroad-spectrum antiviral compound that inhibits the replication of multiple viruses in cell culture (e.g., Chikungunya virus,Venezuela equine encephalitis virus, respiratory syncytial virus,hepatitis C virus, norovirus, influenza A and B viruses, Ebola virus, and human coronaviruses) (3). The triphosphorylated derivative of NHC is a substrate for viral RNA polymerases and interferes with viral replication. In cell culture assays, molnupiravir was found to be a potent inhibitor of SARS-CoV-2replication with an EC50 in the submicromolar range (1,2 ,3, 4).
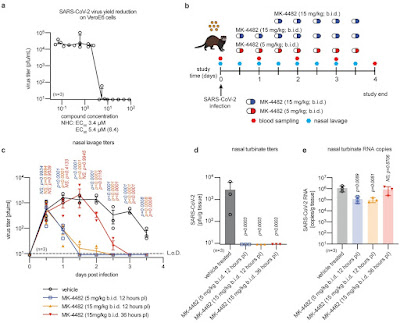
Fig.1 Therapeutic MK-4482/EIDD-2801 is orally efficacious against SARS-CoV-2 in ferrets (8)
Plemper also discovered a new molecule 4 Floro Uridine, with remarkable actions inhibiting SARS CoV 2 RNA polymerase (Fig 2) (8).
Fig. 2 Structure of the analogue nucleoside 4 Fluoro Uridine.
This inhibitory effect was also observed in animal models such as Syrian hamsters and humanized mice (5, 6), and NHC administration to infected ferrets prevented SARS-CoV-2 transmission to untreated and uninfected animals (7). Molnupiravir is currently approved by a narrow margin of 13 to 10 votes (Emergecy Use Approbation) by FDA. Studies carried out in cell culture with alphaviruses and coronaviruses have shown the mutagenic action of NHC, inducing G to A and C to U transitions in a dose dependent manner.
Plemper (8) in the Georgia State University established a ferrets model to show Molnipiravir action in ferrets (Fig. 1 y 3).
Fig. 3. Therapeutic oral treatment with MK-4482/EIDD-2801 prevents contact transmisión (14).
In their recent work, Gordon et al. (9) used a SARS-CoV-2RdRp holoenzyme and various RNA-RNA template primers to determine nucleotide incorporation efficiencies for NHC triphosphate and natural ribonucleoside triphosphates (rNTPs)in different sequence contexts. Although accurate determinations of kpol (polymerase catalytic rate constant) and Kd (nucleotide binding affinity) were not obtained, their results clearly showed that all rNTPs are more efficiently incorporated than the NHC triphosphate. However, as expected, the most efficient competition was observed with cytosine triphosphate(CTP). In a different set of experiments and found that once incorporated, primers having NHC monophosphate at their 3’ end were efficiently extended, particularly at high rNTP concentrations.
This observation led the authors to investigate whether a template NHC could have an impact on the generation of errors during SARS-CoV-2 RNA synthesis. A RNA template containing an embedded NHC monophosphate was synthesized and used in RNA elongation experiments, which demonstrated the formation of both NHC:A and NHC:G base pairs. However, efficient extension was only observed in the case of NHC:A base-pairing. These biochemical data led the authors ( Menéndez-Arias 16. REFERENCIA 10) to propose a model to explain the mutagenic antiviral action of NHC (Fig. 4).
Fig 4. Proposed mechanism of action of Molnupiravir ( 16, 17. REVISAR).While the formation of NHC:G base pairs could lead to RNA synthesis inhibition, NHC:A pairing induced mutagénesis.
Subbstitution of Adenine and Guanine by Molnupiravir is proposed also by (FALTA COMPLETAR ESTE PÁRRAFO)
Fig.5 Proposed MOA of Molnupiravir.(17)
Error catastrophe could be defined as a cumulative loss of genetic information in alineage of organisms due to high mutation rates. The process leading to viral extinction through the accumulation of errors is known as lethal mutagenesis. Error catastrophe occurs when the mutation rate exceeds an error threshold. Viruses and bacteria have evolved to mainto maintain a characteristic error rate. RNA viruses have very high mutation rates and replicate near the error threshold for the maintenance of genetic information [10].
A modest 1.1- to 2.8-fold increase in their mutation frequency can be sufficient to enter error catastrophe, as shown for vesicular stomatitis virus and poliovirus [11] organisms due to high mutation rates. The process leading to viral extinction through the accumulation of errors is known as lethal mutagenesis. Error catastropheoccurs when the mutation rate exceeds an error threshold. Viruses and bacteria haveevolved to maintain a characteristic error rate. RNA viruses have very high mutation ratesand replicate near the error threshold for themaintenance of genetic information [10].Molnupiravir iS a prodrug and must be metabolized cleavage the ester bound (red caps) ,
there is similarity of action with the metabolite GS-441524MP, from Remdesivir.
The term ‘lethal mutagenesis’ was coined in 1999 by Loeb and colleagues after showing that exposing the human immunodeficiency virus type 1 (HIV-1) LAI strain to 1mM 5- hydroxy deoxycytidine (Figure 1) led to the loss of viral titer after 24 sequential passages [13].
The sequencing of part of the HIV-1 reverse transcriptase (RT) coding region of the penultimate passage prior to extinction revealed 2.6- and 5-fold increases in the frequency of A-to-G transitions (A→G) in two separate experiments [13]. These results stimulated further studies addressing the potential of mutagenic nucleoside analogs as antiviral agents driving viral populations to extinction.
Conclusions.
Nucleotide analogues Remdesivir, Molnipiravir and 4 -Florouridine and Paxlovid remains as the only drugs approved by regulatory agencies to treat COVID-19. However, some clinical studies have failed to confirm its beneficial effects specially on Remdesivir. Another disavantages is his high price and i.v. administration. In addition, the drug is difficult to synthesize, expensive, and has to be administered intravenously in a hospital setting. These attributes are undesirable in the contextof a pandemic and efforts have been focused on the development of alternative inhibitors of SARS-CoV-2 replication.
On the contrary, Molnipiravir and Paxlovid are oral drugs , witha good bioavailability . Among them, Molnupiravir has emerged as a promising new at dose of 800 mg b.i.d. for 5 days (price 530 dollar). Paxolvid is also useful (price of treatment 730 dollars).
A new drug such as 4-Florourudine, also an analogue of Uracil, has a “terminal chain effect” that must be carefully studied.
References
1. Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [CrossRef]
[PubMed]
2. Domingo, E.; García-Crespo, C.; Perales, C. Historical perspective on the discovery of the quasispecies concept. Annu. Rev. Virol.
2021, 8, 51–72.
3. Eigen, M. Error catastrophe and antiviral strategy. Proc. Natl. Acad. Sci. USA 2002, 99, 13374–13376.
4. Eigen, M. From Strange Simplicity to Complex Familiarity: A Treatise on Matter, Information, Life and Thought; Oxford University Press:
Cary, NC, USA, 2013.
5. Weissmann, C.; Billeter, M.A.; Goodman, H.M.; Hindley, J.; Weber, H. Structure and function of phage RNA. Annu. Rev. Biochem.
1973, 42, 303–328.
6. Haruna, I.; Spiegelman, S. Recognition of size and sequence by an RNA replicase. Proc. Natl. Acad. Sci. USA 1965, 54, 1189–1193.
7. Haruna, I.; Spiegelman, S. Specific template requirments of RNA replicases. Proc. Natl. Acad. Sci. USA 1965, 54, 579–587.
8. Drake, J.W.; Charlesworth, B.; Charlesworth, D.; Crow, J.F. Rates of spontaneous mutation. Genetics 1998, 148, 1667–1686.
9. Drake, J.W.; Holland, J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 1999, 96, 13910–13913. [CrossRef]
10. Perales, C.; Domingo, E. Antiviral strategies based on lethal mutagenesis and error threshold. Curr. Top. Microbiol. Immunol. 2016,
392, 323–339.
11. Holland, J.J.; Domingo, E.; de la Torre, J.C.; Steinhauer, D.A. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 1990, 64, 3960–3962.
12. Anderson, J.P.; Daifuku, R.; Loeb, L.A. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 2004, 58, 183–205.
13. Loeb, L.A.; Essigmann, J.M.; Kazazi, F.; Zhang, J.; Rose, K.D.; Mullins, J.I. Lethal mutagenesis of HIV with mutagenic nucleosideanalogs. Proc. Natl. Acad. Sci. USA 1999, 96, 1492–1497.
(14) . Cox RM, Wolf JD, Plemper RK. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat Microbiol. 2021 Jan;6(1):11-18. doi: 10.1038/s41564-020-00835-2. Epub 2020 Dec 3. PMID: 33273742; PMCID: PMC7755744.
(15) Kabinger F, Stiller C, Schmitzová J, Dienemann C, Kokic G, Hillen HS, Höbartner C, Cramer P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat Struct Mol Biol. 2021 Sep;28(9):740-746. doi: 10.1038/s41594-021-00651-0. Epub 2021 Aug 11. PMID: 34381216; PMCID: PMC8437801.
(16) Hadj Hassine I, Ben M'hadheb M, Menéndez-Arias L. Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity. Viruses. 2022 Apr 18;14(4):841. doi: 10.3390/v14040841. PMID: 35458571; PMCID: PMC9024455.
(17) Menéndez-Arias L. Decoding molnupiravir-induced mutagenesis in SARS-CoV-2. J Biol Chem. 2021 Jul;297(1):100867. doi: 10.1016/j.jbc.2021.100867. Epub 2021 Jun 9. PMID: 34118236; PMCID: PMC8188802.